Day 24 Split plot designs
July 15th, 2025
24.2 Split plot designs
- Split-plot designs are multi-level designed experiments.
- Randomization happens in different domains.
Let’s look at two alternatives for randomizing the whole-plot factor levels, in a CRD and in a blocked scenario:
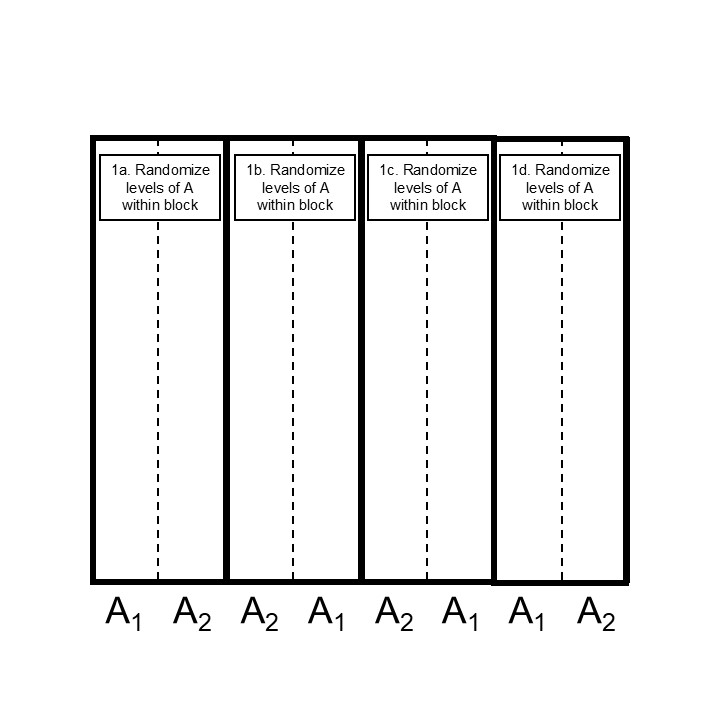
Figure 24.1: Step 2 designing a split plot design
Remember:
- Blocks are groups of approximately similar experimental units.
- Data generated by blocked designs may be analyzed as \(y_{ij} = \mu +\tau_i + b_j + \varepsilon_{ij}\), where \(y_{ij}\) is the observed response for the \(i\)th treatment in the \(j\)th block, \(\tau_i\) is the effect of the \(i\)th treatment, \(b_j\) is the effect of the \(j\)th block, \(\varepsilon_{ij}\) is the residual of the \(i\)th treatment in the \(j\)th block.
- Data generated by split-plot designs may be analyzed as \(y_{ijk} = \mu + T_i +A_j +(TA)_{ij} + b_k + w_{i(k)} + \varepsilon_{ij}\), where \(y_{ij}\) is the observed response for the \(i\)th level of factor T, the \(j\)th level of factor A, in the \(k\)th block, \(T_i\) is the effect of the \(i\)th level of treatment T, \(A_j\) is the effect of the \(j\)th level of treatment A, \(b_k\) is the effect of block \(k\) and may be random or fixed, \(w_i(k)\) is the effect of the whole plot corresponding to the \(i\)th treatment T in block \(k\), and \(\varepsilon_{ijk}\) is the residual of the \(i\)th level of factor T, the \(j\)th level of factor A, in the \(k\)th block.
- Typically assume \(w_{i(k)} \sim N(0, \sigma^2_w)\).
- \(b_k\) may still be fixed or random.
- Variance components should be \(> 0\).
24.2.1 Applied case: split-plot design
Grain yield was measured at for a split-plot experiment of barley with fungicide treatments. Within each block, fungicides were randomized and then, barley varieties were randomly applied to smaller plots.
library(tidyverse)
library(agridat)
library(lme4)
library(emmeans)
df_splitplot <- agridat::durban.splitplot
m_splitplot <- lmer(yield ~ fung * gen + (1|block/fung),
data = df_splitplot)
VarCorr(m_splitplot)## Groups Name Std.Dev.
## fung:block (Intercept) 0.11737
## block (Intercept) 0.16972
## Residual 0.2811924.2.1.1 Standard errors
sigma2_e <- sigma(m_splitplot)^2
sigma2_wp <- as.data.frame(VarCorr(m_splitplot))[1,]$vcov
sigma2_b <- as.data.frame(VarCorr(m_splitplot))[2,]$vcov
levels_wp <- n_distinct(df_splitplot$fung)
levels_sp <- n_distinct(df_splitplot$gen)
reps <- n_distinct(df_splitplot$block)Mean differences for factor at the whole plot
## [1] 0.086331## NOTE: Results may be misleading due to involvement in interactions## contrast estimate SE df t.ratio p.value
## c(1, -1) 0.548 0.0863 3 6.346 0.0079
##
## Results are averaged over the levels of: gen
## Degrees-of-freedom method: kenward-rogerMean differences for factor at the sub plot
## [1] 0.1405927## NOTE: Results may be misleading due to involvement in interactions## contrast estimate SE df t.ratio p.value
## c(1, -1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, -0.152 0.141 414 -1.085 0.2787
##
## Results are averaged over the levels of: fung
## Degrees-of-freedom method: kenward-roger